We Out Here Tryin' to Function
Aug. 2, 2021
I think one of the key steps in making “improvements” on your R journey is to write code that is more clear, concise and short; for me, this usually occurs when I need to create multiple plots of variables when exploring a dataset.
I find myself usually making simple bar plots using ggplot and geom_col() to count things. Instead of copying-and-pasting the same ggplot code and altering the the column names in the code like a newbie hack, I recently learned a new function that can handle the tidyverse workflow when making bar plots.
First, we’ll use the palmerpenguins package to make some bar plots (Yeah, I am fully aware that I should’ve worked with some Spotify/Apple Music Bay Area music dataset in order to make this blog post even more hyphy…). I examine the penguins dataset to see which columns are categorical/discrete and numerical/continuous:
library(tidyverse)
library(palmerpenguins)
theme_set(theme_minimal())
penguins %>% glimpse()
## Rows: 344
## Columns: 8
## $ species <fct> Adelie, Adelie, Adelie, Adelie, Adelie, Adelie, Adel…
## $ island <fct> Torgersen, Torgersen, Torgersen, Torgersen, Torgerse…
## $ bill_length_mm <dbl> 39.1, 39.5, 40.3, NA, 36.7, 39.3, 38.9, 39.2, 34.1, …
## $ bill_depth_mm <dbl> 18.7, 17.4, 18.0, NA, 19.3, 20.6, 17.8, 19.6, 18.1, …
## $ flipper_length_mm <int> 181, 186, 195, NA, 193, 190, 181, 195, 193, 190, 186…
## $ body_mass_g <int> 3750, 3800, 3250, NA, 3450, 3650, 3625, 4675, 3475, …
## $ sex <fct> male, female, female, NA, female, male, female, male…
## $ year <int> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007…
An easy first look at exploring a dataset is to simply count the number of items in a variable. I want to determine the counts across the categorical columns and make a bar plot for each. That happens to be the species, island and sex columns. Here’s the long way how to do it:
penguins %>%
drop_na() %>%
count(species, sort = TRUE) %>%
mutate(species = fct_reorder(species, n)) %>%
ggplot(aes(x = species, y = n)) +
geom_col() +
coord_flip()
penguins %>%
drop_na() %>%
count(island, sort = TRUE) %>%
mutate(island = fct_reorder(island, n)) %>%
ggplot(aes(x = island, y = n)) +
geom_col() +
coord_flip()
penguins %>%
drop_na() %>%
count(sex, sort = TRUE) %>%
mutate(sex = fct_reorder(sex, n)) %>%
ggplot(aes(x = sex, y = n)) +
geom_col() +
coord_flip()
(There should be three plots but I’ve disabled the output for brevity’s sake). In the code demonstrated above, I realize that all I am just changing is the name of the columns (species, island, sex) to create the three different plots. The rule of thumb is to avoid duplication of code; here we can attempt to create a function to shorten the number of lines written. We can create a function (I named it geomcol_discrete) with the nifty use of double brackets {{ column }} to maintain a tidyverse work flow:
# function - discrete plots
geomcol_discrete <- function(tbl, column) {
tbl %>%
drop_na() %>%
count({{ column }}, sort = TRUE) %>%
mutate({{ column }} := fct_reorder({{ column }}, n)) %>%
ggplot(aes(x = {{ column }}, y = n)) +
geom_col() +
coord_flip()
}
# now we quickly plot
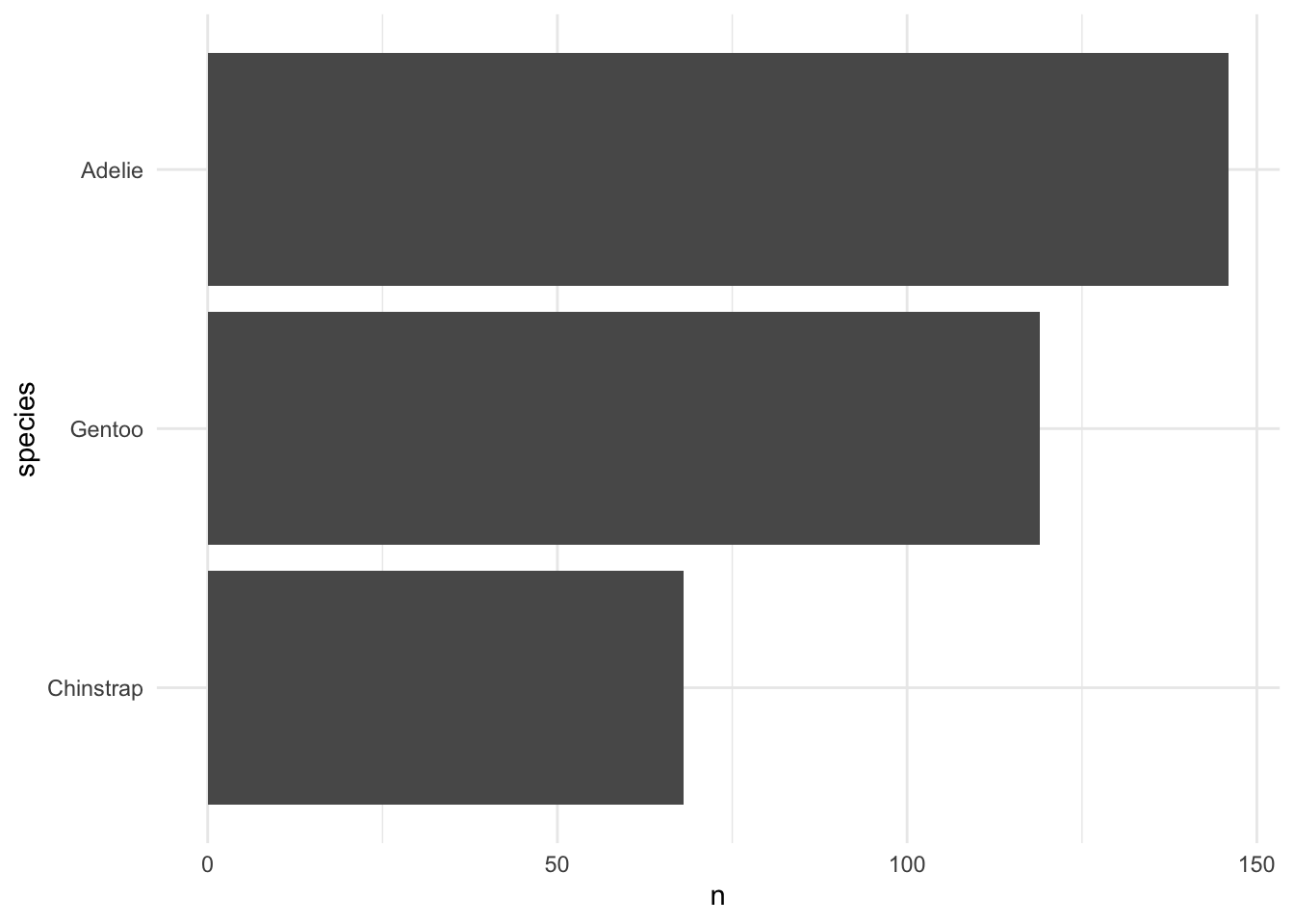
penguins %>% geomcol_discrete(species)
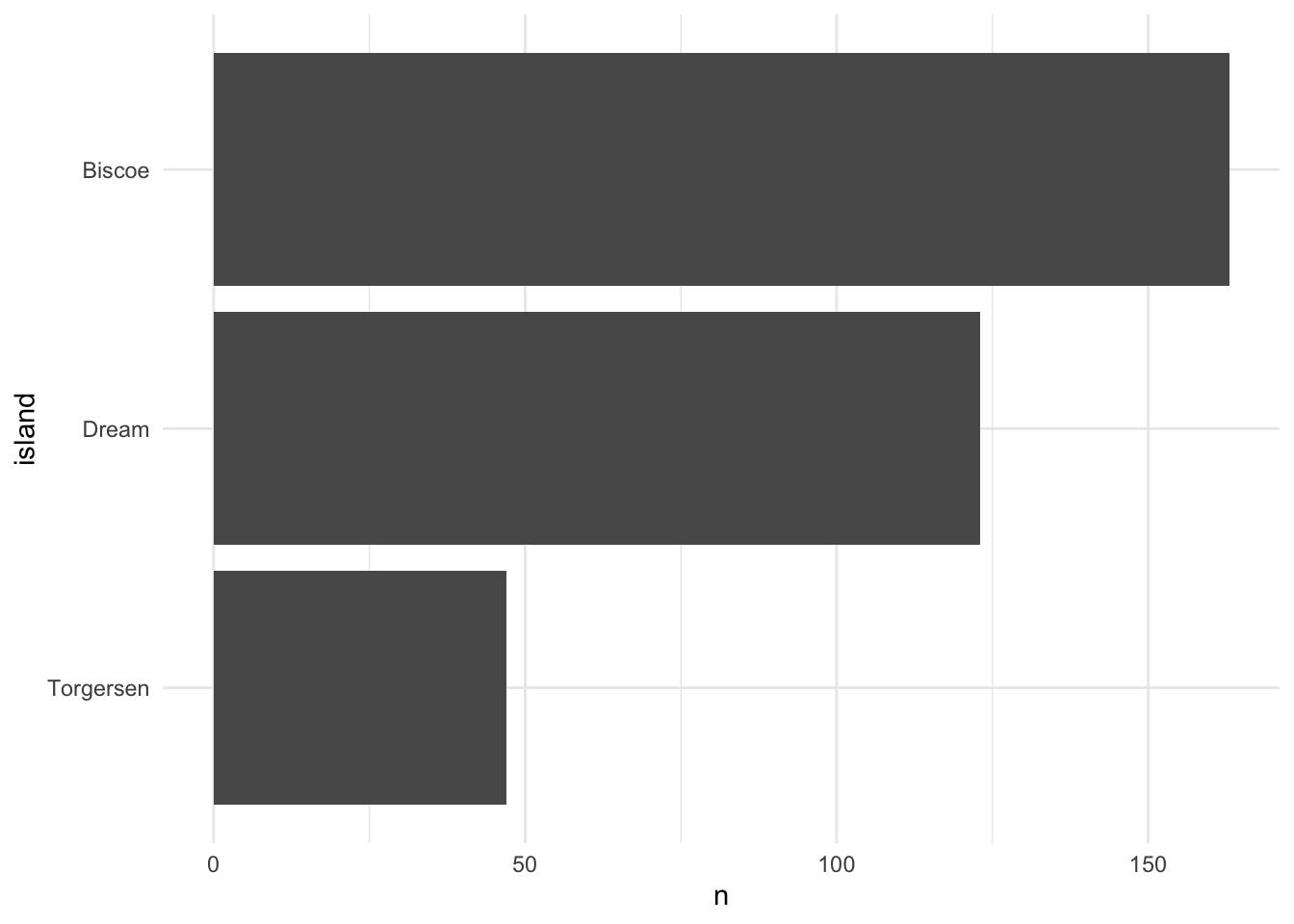
penguins %>% geomcol_discrete(island)
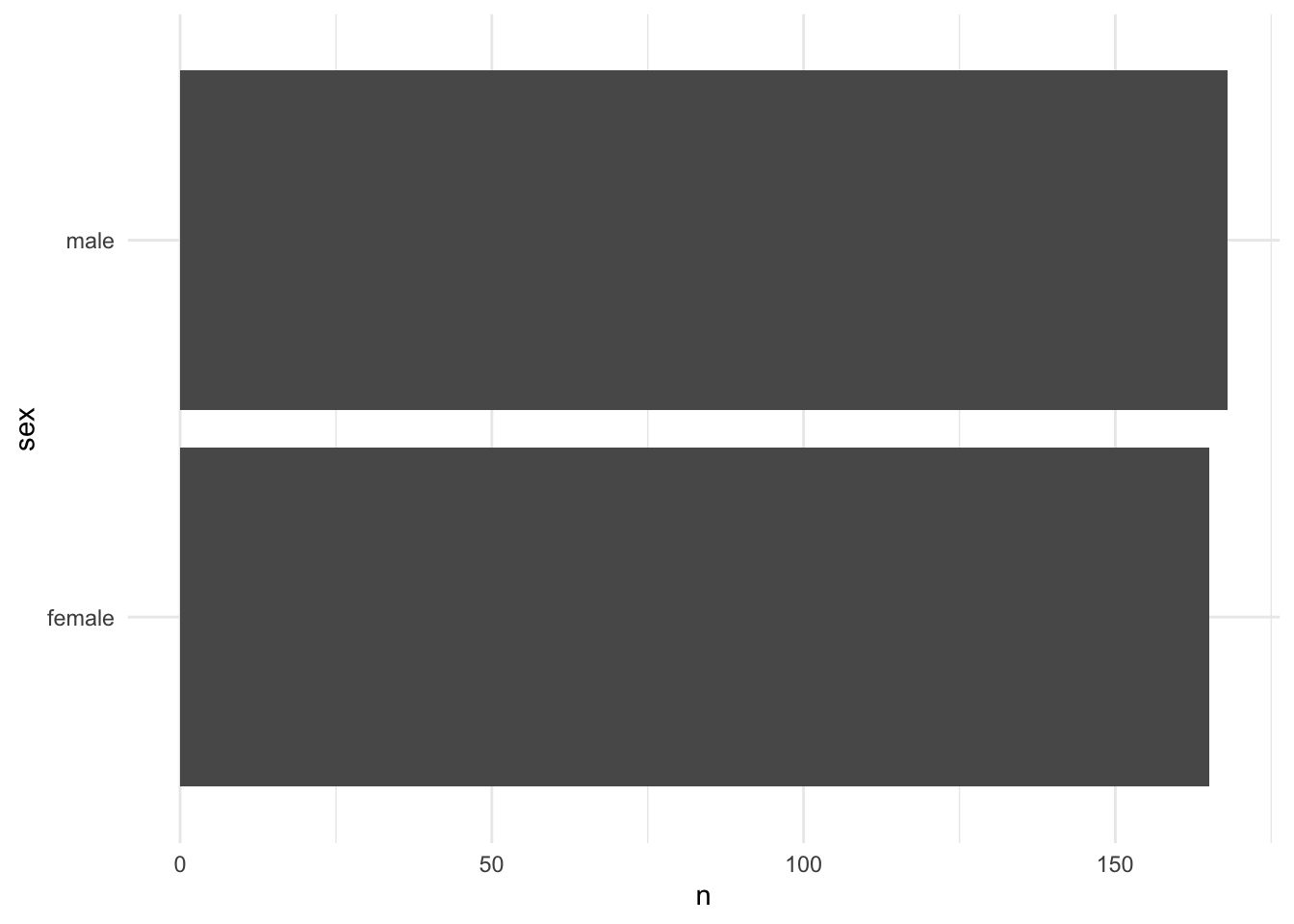
penguins %>% geomcol_discrete(sex)
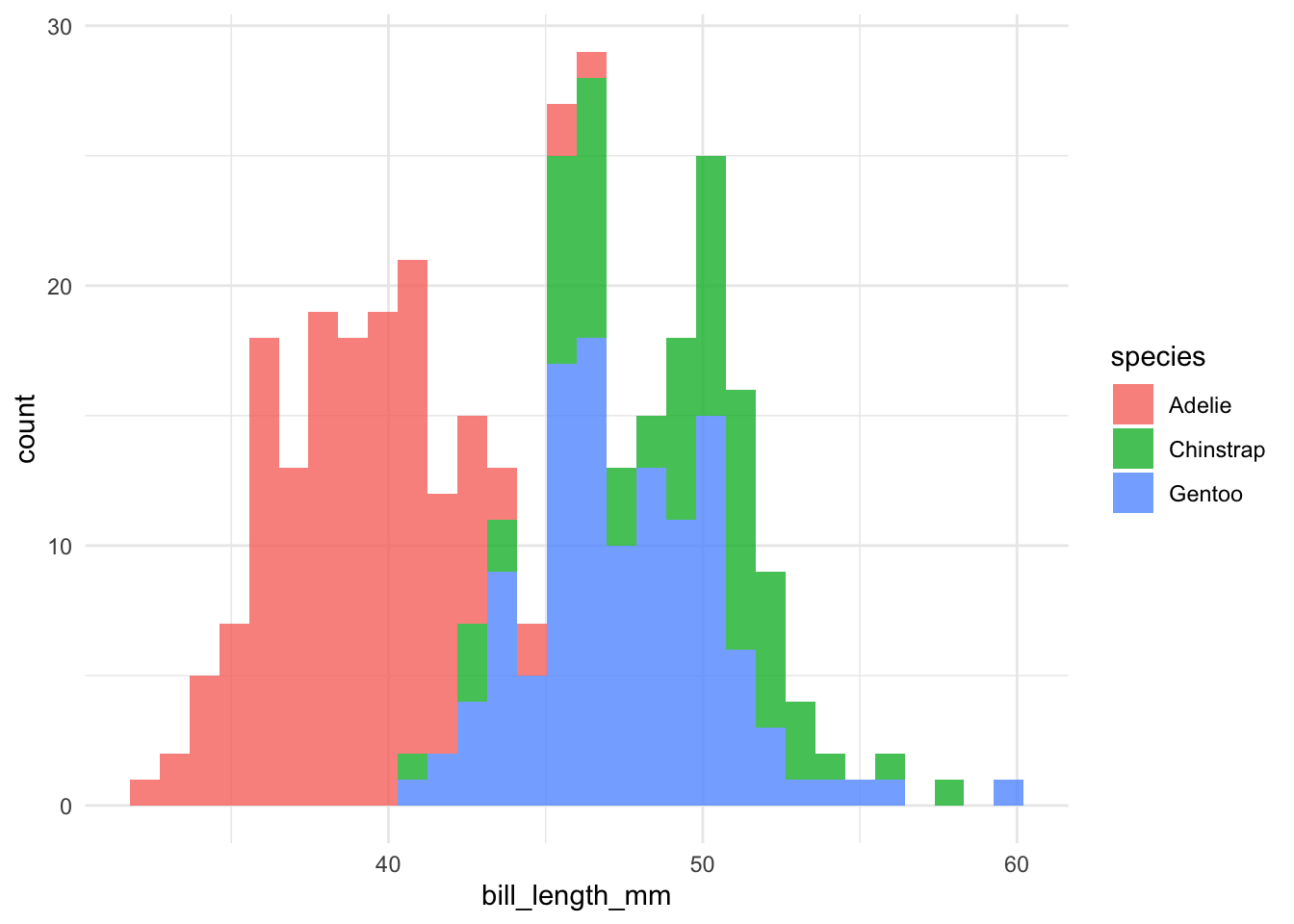
I also created a function to plot histograms for the continuous data (geomhist_continuous):
# function - continuous plots
geomhist_continuous <- function(tbl, column) {
tbl %>%
drop_na() %>%
ggplot(aes(x = {{ column }}, fill = species)) +
geom_histogram(alpha = 0.8)
}
# now we quickly plot
penguins %>% geomhist_continuous(bill_length_mm)
penguins %>% geomhist_continuous(bill_depth_mm)
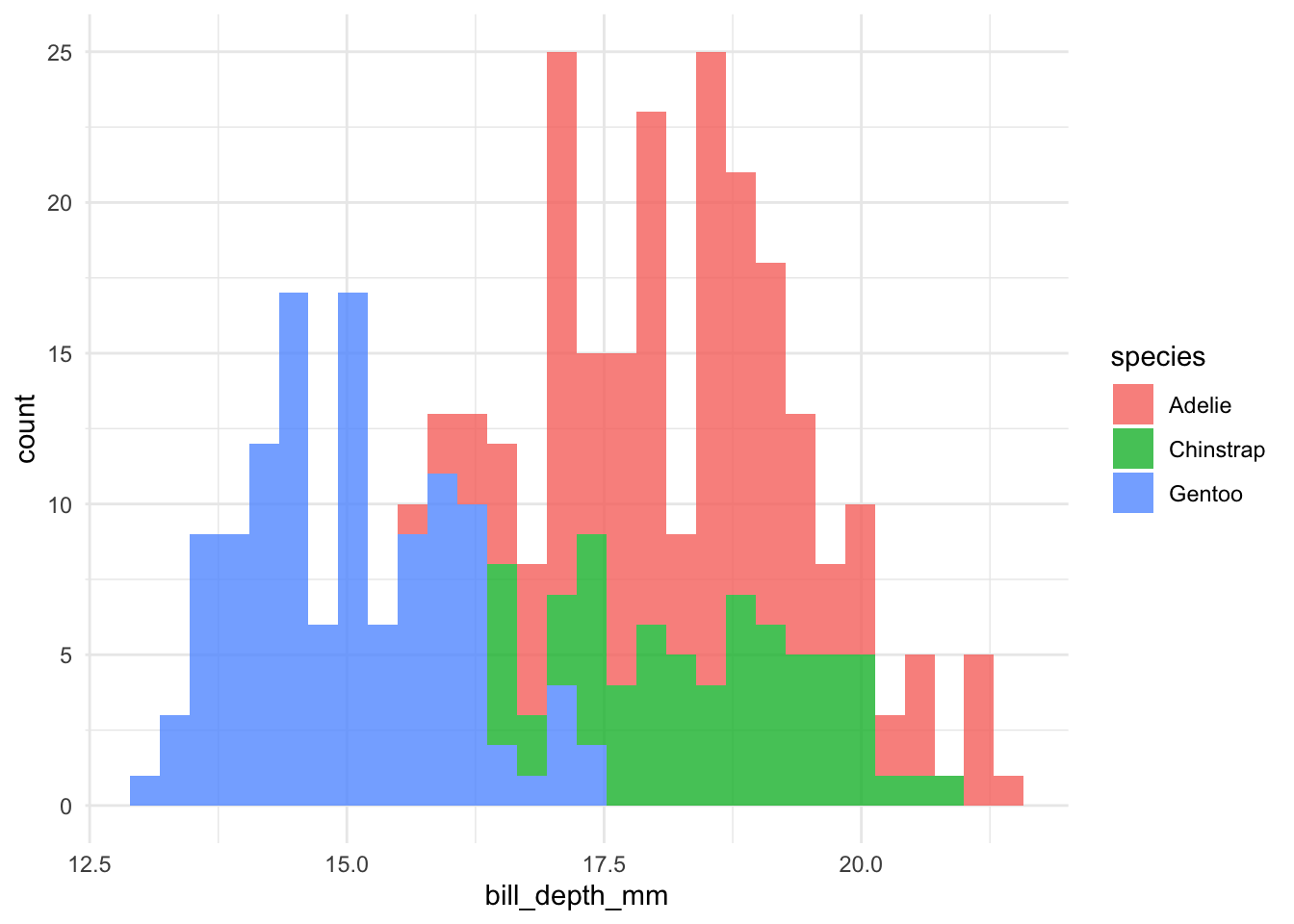
penguins %>% geomhist_continuous(flipper_length_mm)
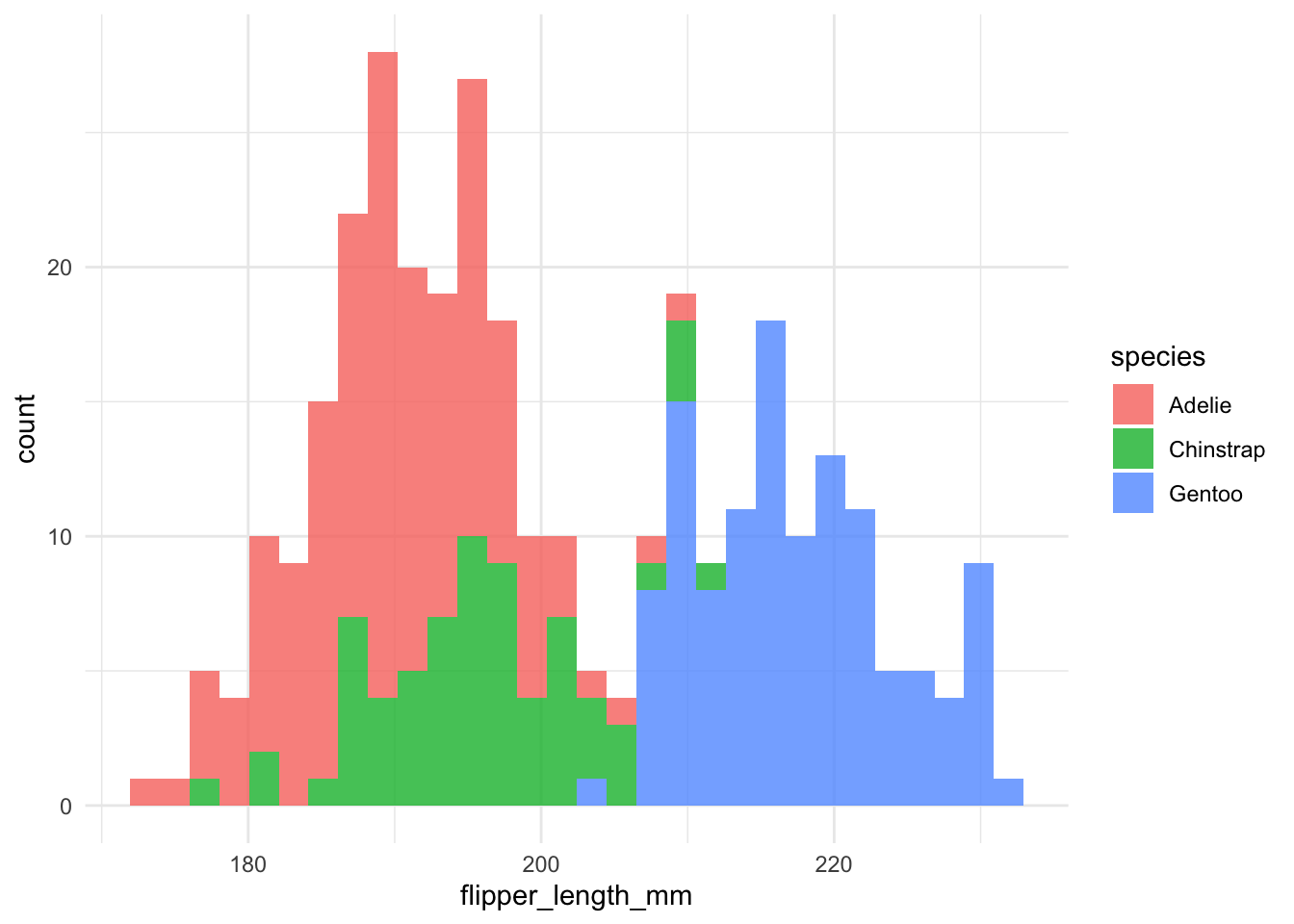
penguins %>% geomhist_continuous(body_mass_g)
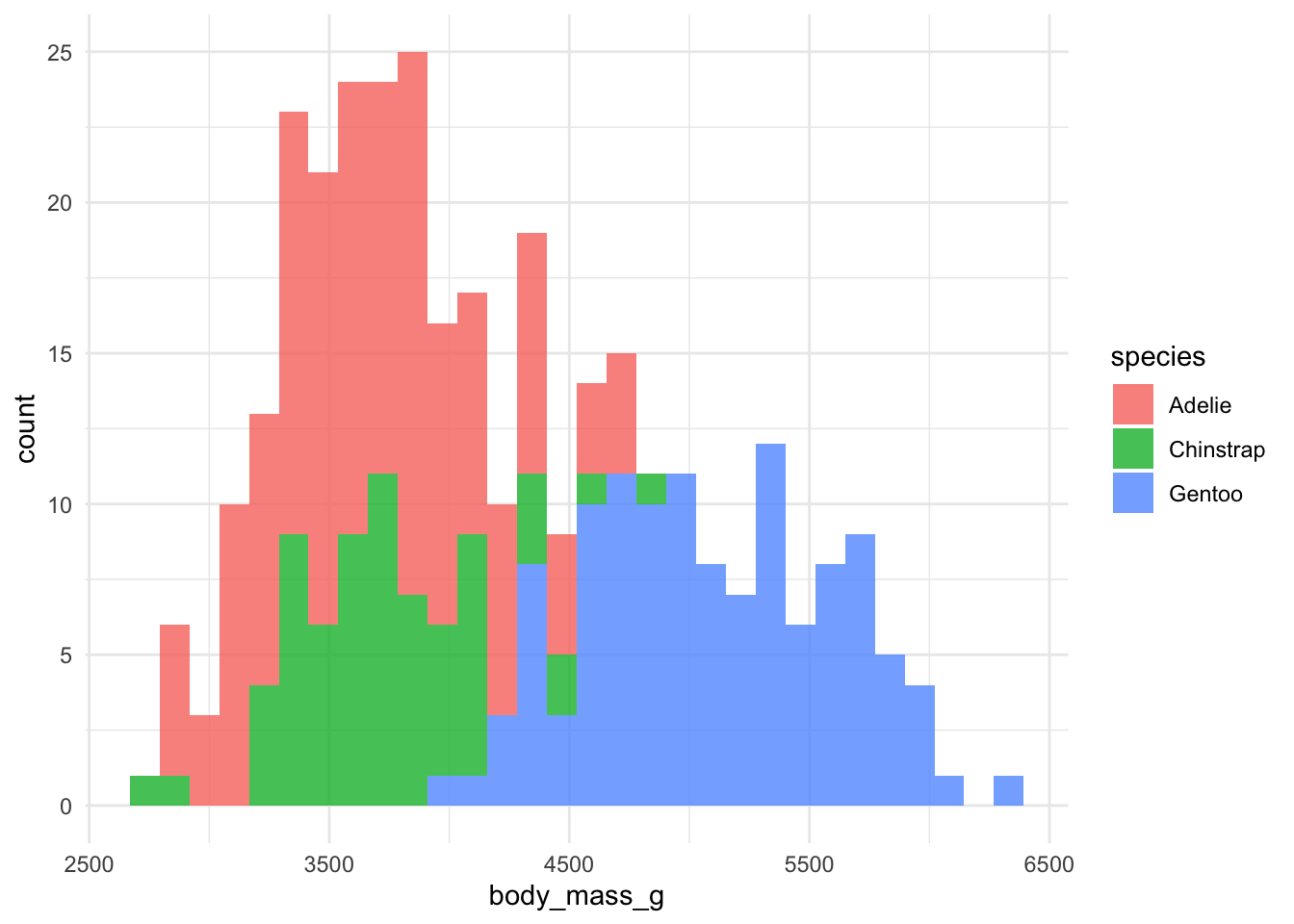
The next step further would be to create a vector of column names so that I can loop the functions. Stay tuned for a future update. As the Bay Area colloquially says, “We out here tryin’ to function!”